Junk DNA
- 10 Nov 2025
In News:
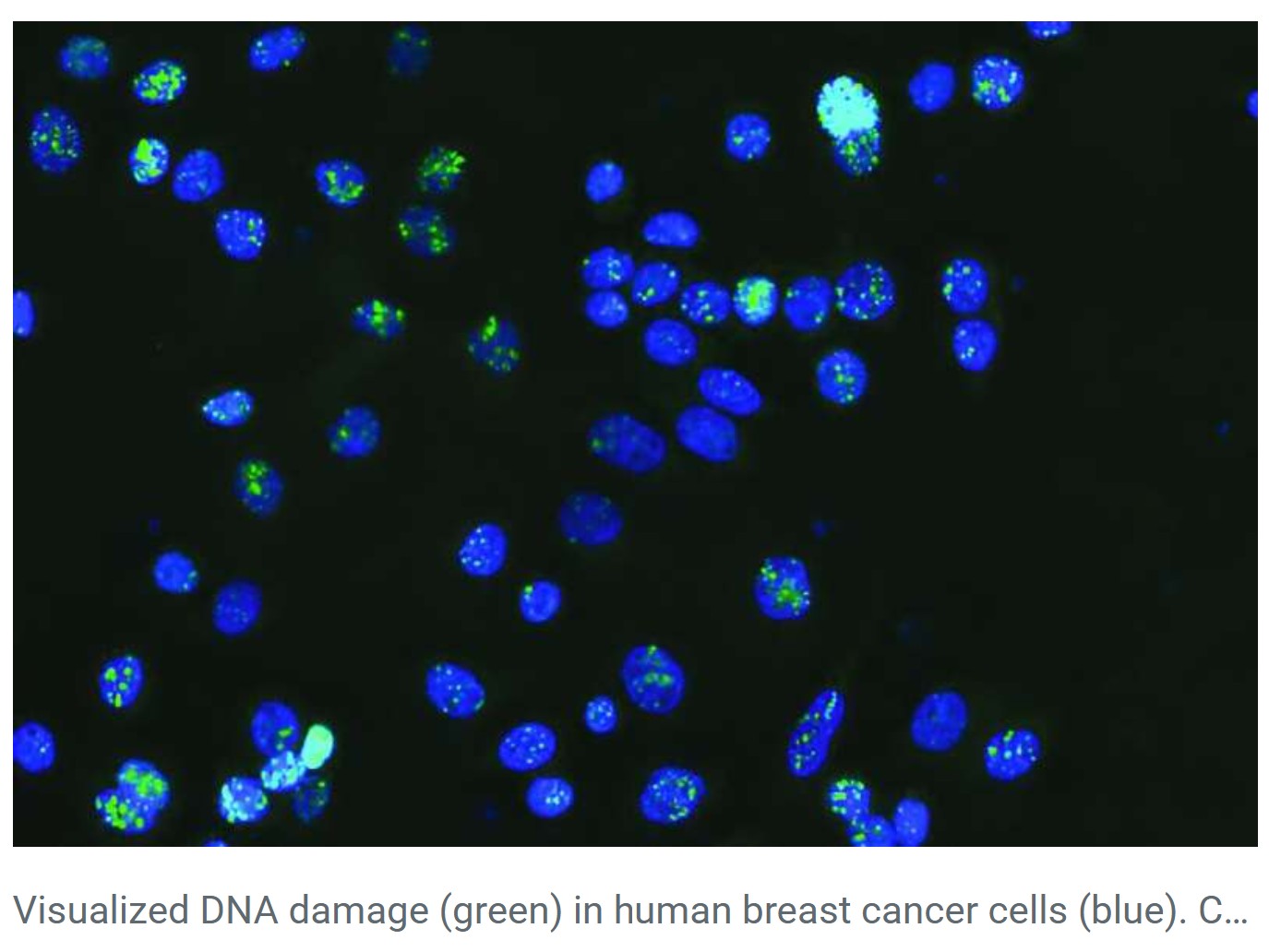
For decades, large portions of the human genome were dismissed as “junk DNA”- genetic material assumed to have little or no functional value. However, advances in genetics, genomics, and artificial intelligence are rapidly reshaping this understanding. Recent research, particularly the discovery of cancer-linked mutations in non-coding DNA, highlights how so-called junk DNA plays a crucial role in gene regulation, genome architecture, evolution, and disease.
Understanding Junk DNA
In genetics, junk DNA refers to regions of DNA that do not code for proteins. While DNA’s primary role is to provide instructions for protein synthesis, not all DNA sequences serve this function.
- In the human genome, nearly 98% of DNA is non-coding, whereas in simpler organisms like bacteria, only about 2% of DNA is non-coding.
- A part of non-coding DNA is known to have clear functions, such as producing:
- Transfer RNA (tRNA)
- Ribosomal RNA (rRNA)
- Regulatory RNAs
- However, a substantial fraction neither codes for proteins nor produces RNA, and its function remained unclear- hence the term junk DNA.
Over time, scientists have accumulated evidence that these regions are not entirely useless. Some DNA fragments that were originally non-functional have acquired functions through exaptation—a process by which structures or sequences evolve new roles not originally shaped by natural selection.
Emerging Functional Significance of Non-Coding DNA
Modern genomics has revealed that non-coding DNA plays a vital role in:
- Gene regulation (switching genes on or off),
- Chromatin organisation,
- Genome stability,
- Evolutionary innovation.
These roles become especially critical in understanding complex diseases such as cancer, where gene regulation and genome structure are often disrupted.
Breakthrough Discovery: Cancer Mutations in ‘Junk’ DNA
A recent study by the Garvan Institute of Medical Research, published in Nucleic Acids Research, marks a major breakthrough. Using artificial intelligence and machine learning, researchers identified a new class of cancer-driving mutations hidden in non-coding DNA.
Key Findings
- Mutations were found in non-coding regions across at least 12 cancer types, including breast, prostate, and colorectal cancers.
- Every tumour sample analysed had at least one mutation in these critical non-coding regions.
- These mutations were located at specific DNA sites that bind a protein called CTCF (CCCTC-binding factor).
Role of CTCF and Genome Architecture
CTCF is a key protein that helps fold long strands of DNA into precise three-dimensional (3D) structures inside the nucleus. These structures act as genomic “anchors”, bringing distant DNA regions together and controlling which genes are expressed.
- Some CTCF binding sites are “persistent anchors”, meaning they are present across many cell types.
- Mutations at these sites disrupt the 3D organisation of the genome, leading to abnormal gene activation or suppression.
- Such disruptions give cancer cells a survival and growth advantage, turning these sites into mutational hotspots.
To identify these sites, researchers developed an AI-based tool called CTCF-INSITE, which analysed genomic and epigenomic data from over 3,000 tumour samples using data from the International Genome Consortium.
Implications for Cancer Diagnosis and Treatment
This discovery has far-reaching implications:
- Universal cancer targets: Since the same non-coding mutations appear across multiple cancers, therapies could potentially work across cancer types rather than being mutation-specific.
- Early diagnosis: Alterations in these genomic anchors could serve as biomarkers for early cancer detection.
- New treatment strategies: Researchers plan to use CRISPR gene-editing to study how correcting these mutations affects cancer progression.
- AI in healthcare: The study demonstrates how artificial intelligence can uncover hidden patterns in vast biological data sets.
DNA Identification in Mass Fatality Events
- 17 Jun 2025
In News:
Following the tragic crash of an Air India Boeing 787 Dreamliner from Ahmedabad to London Gatwick (June 2025), authorities have initiated DNA-based identification to match the remains of victims. In mass fatality incidents where bodies are mutilated or decomposed, DNA analysis becomes the gold standard for establishing identity.
What is DNA Identification?
DNA (Deoxyribonucleic Acid) is a unique genetic code present in almost every cell of the human body, with the exception of identical twins. It is widely used in forensic science for accurate identification, particularly in disasters where visual identification is impossible.
Sample Collection and Preservation:
- DNA begins degrading post-mortem, and the rate of degradation is influenced by:
- Type of tissue (soft vs hard)
- Environmental conditions (humidity, temperature)
- Hard tissues such as bones and teeth are preferred due to better preservation against decomposition.
- Soft tissues (like skin and muscle) degrade faster and, if used, must be stored in 95% ethanol or frozen at -20°C.
- In large-scale accidents, sample collection from wreckage can take weeks or even months (e.g., 9/11 took 10 months).
Reference Samples:
To match unidentified remains, reference DNA is taken from biological relatives—preferably parents or children of the victims, who share about 50% of their DNA.
Methods of DNA Analysis:
1. Short Tandem Repeat (STR) Analysis:
- Evaluates short, repeating DNA sequences that vary among individuals.
- Requires nuclear DNA, hence not suitable if the DNA is highly degraded.
- Analysis of 15+ hyper-variable STR regions can confirm family relationships with high accuracy.
2. Mitochondrial DNA (mtDNA) Analysis:
- Used when nuclear DNA is not recoverable.
- mtDNA is inherited exclusively from the mother and is present in multiple copies per cell.
- Effective for matching with maternal relatives (e.g., mother, maternal uncles/aunts, siblings).
3. Y-Chromosome Analysis:
- Targets male-specific genetic material.
- Useful for identifying remains using DNA from paternal male relatives (father, brothers, paternal uncles).
- Helpful when direct relatives are unavailable but male-line relatives exist.
4. Single Nucleotide Polymorphisms (SNPs) Analysis:
- Suitable when DNA is highly degraded.
- Analyzes variations at single base-pair locations in DNA.
- Can also match DNA with personal items like a toothbrush or hairbrush.
- However, less accurate than STR analysis.
Significance for Disaster Management and Forensics:
- DNA-based victim identification ensures scientific accuracy, aiding in closure for families, and upholding legal and humanitarian obligations.
- Modern forensic genetics has become an essential tool in mass disaster response protocols worldwide.
Next Generation DNA Sequencing Facility (NGS)

- 22 Dec 2024
Recently, the Union Minister Shri Bhupender Yadav inaugurated two groundbreaking facilities at the Wildlife Institute of India (WII), Dehradun: the Advanced Facility for Pashmina Certification and the Next Generation DNA Sequencing (NGS) Facility. These facilities are designed to enhance India’s capabilities in wildlife conservation and support the growth of traditional crafts like Pashmina weaving.
Key Highlights
Next Generation DNA Sequencing Facility (NGS)
The NGS facility is a cutting-edge research tool that enables the high-throughput analysis of entire genomes. This technology is pivotal in studying wildlife genetics and biodiversity by decoding millions of DNA sequences at once.
Applications in Wildlife Conservation:
- Genetic Diversity and Health: NGS helps assess the genetic diversity of species and their population health.
- Evolutionary Relationships: It aids in understanding the evolutionary history and unique adaptations of species.
- Disease Surveillance: The technology supports studying pathogen-host interactions and monitoring diseases affecting wildlife.
- Combating Illegal Wildlife Trade: NGS can help detect illegal wildlife trade and the movement of endangered species.
- Impact of Climate Change: It is crucial for studying how climate change affects genetic diversity and species survival.
This facility positions WII as a leading hub for molecular research, enabling more precise conservation efforts and studies on endangered species like tigers, elephants, and riverine dolphins.
Advanced Facility for Pashmina Certification
Launched under a Public-Private Partnership (PPP) model between WII and the Export Promotion Council for Handicrafts (EPCH), the Pashmina Certification Centre (PCC) has been significantly upgraded. The facility now includes a Scanning Electron Microscope (SEM) with Energy Dispersive Spectroscopy (EDS) for advanced wool testing and certification.
Key Features of the Upgraded Facility:
- Fiber Analysis: The SEM-EDS technology ensures accurate identification and certification of Pashmina fibers, free from any prohibited materials.
- Unique ID and E-certificates: Each certified product is tagged with a unique ID and e-certificate, enhancing traceability and authenticity.
- Global Trade Facilitation: The certification process eliminates delays at exit points, ensuring smoother international trade for certified Pashmina products.
The PCC has already certified over 15,000 Pashmina shawls and plays a crucial role in supporting the livelihoods of artisans and weavers in Jammu & Kashmir. By ensuring the authenticity of Pashmina, the facility also helps combat the illegal trade of Shahtoosh wool, which is harmful to the Tibetan antelope (Chiru).
Significance for Artisans and Conservation:
- Support for Artisans: The upgraded facility helps increase the credibility of Pashmina products in global markets, benefiting local artisans and weavers.
- Conservation Impact: By certifying genuine Pashmina products, the initiative indirectly contributes to the conservation of the Tibetan antelope by reducing illegal poaching and trade.
- Sustainability: The PCC is a self-sustaining model that not only supports conservation but also generates revenue and creates job opportunities.
Overview of the Genome India Project
The Genome India Project is a gene mapping initiative launched by the Department of Biotechnology, aiming to create a comprehensive database of genetic variations across the Indian population. The project focuses on understanding genetic diversity and its implications for health, agriculture, and biodiversity conservation in India.
Goals:
- Comprehensive Gene Mapping: The project seeks to map the genetic variations found within India’s diverse population, enabling better healthcare and disease management.
- Conservation and Biodiversity: Insights from the project will also aid in wildlife conservation by understanding the genetic health of endangered species and their ability to adapt to environmental changes.
This initiative is aligned with India’s broader goals of using advanced technologies to address modern conservation challenges and foster a sustainable future.
Kawasaki Disease
- 08 Dec 2024
In News:
Comedian Munawar Faruqui recently opened up about a tough time in his life when his young son was diagnosed with Kawasaki disease.
What is Kawasaki Disease?
- Kawasaki disease is a rare condition that primarily affects children under the age of five.
- It causes inflammation in the blood vessels, including those that supply blood to the heart.
- With early treatment, most children recover without long-term health issues.
Possible Causes:
- The exact cause of Kawasaki disease is not well understood.
- Experts believe it may be triggered by a combination of genetic and environmental factors, including certain infections.
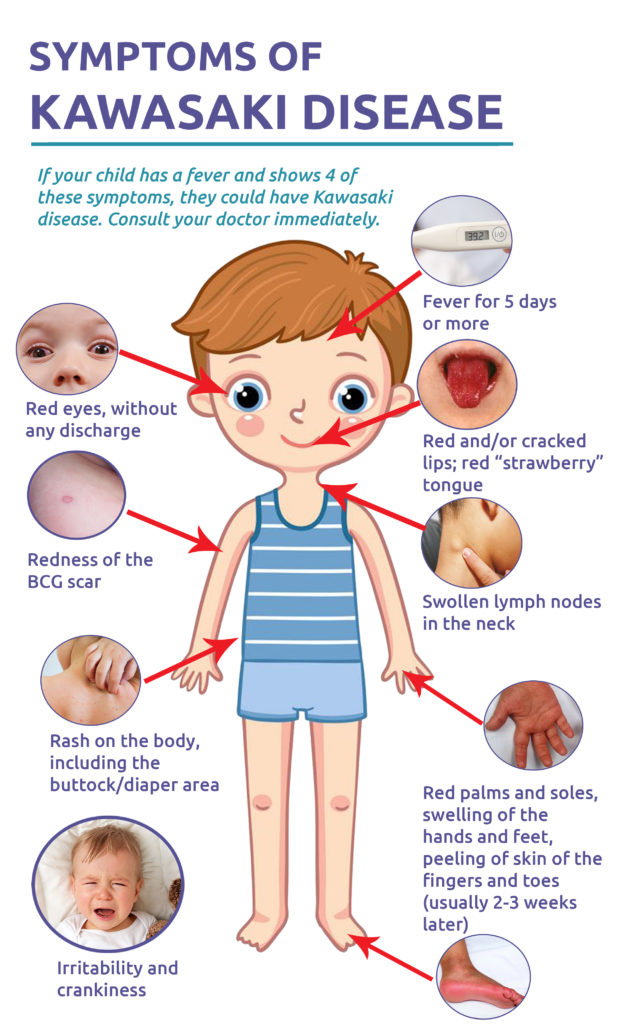
Symptoms: Kawasaki disease symptoms typically appear in two phases and may last for several weeks. Common symptoms include:
- High fever lasting more than five days.
- Red eyes without discharge.
- A rash on the body, particularly in the chest and groin area.
- Swollen hands and feet, sometimes accompanied by redness.
- Red, cracked lips and a swollen, red tongue.
- Swollen lymph nodes, particularly on one side of the neck.
Detection & Treatment:
- There’s no test that can directly detect Kawasaki disease. But healthcare providers can do tests that support a diagnosis of Kawasaki disease or rule out other possible illnesses.
- Treatment for Kawasaki disease includes:Immune globulin (IVIG), or human blood proteins you receive by IV. About 10% of children may not respond to the first dose of IVIG and will need a second dose or other medications.
AlphaFold 3

- 09 May 2024
Why is it in the News?
Google Deepmind has unveiled the third major version of its “AlphaFold” artificial intelligence model, designed to help scientists design drugs and target diseases more effectively.
About AlphaFold 3:
- AlphaFold 3 is a major advancement in artificial intelligence created by Google's DeepMind in collaboration with Isomorphic Labs.
- It's essentially a powerful tool that can predict the structures and interactions of various biological molecules such as:
- Predict structures of biomolecules: Unlike previous versions that focused on proteins, AlphaFold 3 can predict the 3D structure of a wide range of molecules, including DNA, RNA, and even small molecules like drugs (ligands).
- This is a significant leap in understanding how these molecules function.
- Model molecular interactions: AlphaFold 3 goes beyond just structure prediction.
- It can also model how these molecules interact with each other, providing valuable insights into cellular processes and disease mechanisms.
The potential applications of AlphaFold 3 are vast. It has the potential to revolutionize fields like:
- Drug discovery: By understanding how drugs interact with their targets, researchers can design more effective medications.
- Genomics research: AlphaFold 3 can help scientists understand the function of genes and how mutations can lead to disease.
- Materials science: By modelling the interactions between molecules, scientists can design new materials with specific properties.
- AlphaFold 3 is a significant breakthrough and is freely available for non-commercial use through AlphaFold Server.
- This makes this powerful tool accessible to researchers around the world, potentially accelerating scientific advancements.
Attenborough's Long-beaked Echidna (The Hindu)

- 11 Nov 2023
Why in the News?
An elusive echidna feared extinct after disappearing for six decades has been rediscovered in a remote part of Indonesia, on an expedition that also found a new kind of tree-dwelling shrimp.
About Attenborough's long-beaked echidna:
- Attenborough's long-beaked echidna (Zaglossus attenboroughi), also known as Sir David's long-beaked echidna or the Cyclops long-beaked echidna.
- It is an egg-laying mammal native to the Cyclops Mountains in the northern Indonesian region of Papua.
- It is one of three species of long-beaked echidna, and is the smallest and most threatened of the three.
- The echidna was named after naturalist Sir David Attenborough.
- The echidna is a nocturnal animal, and it is most active at night.
- It spends its days sleeping in burrows, and it emerges at night to forage for food.
- The echidna's diet consists of ants, termites, beetles, and other insects.
- Attenborough's long-beaked echidna is classified as Critically Endangered by the IUCN.
Cell-free DNA (The Hindu)
- 01 Aug 2023
Why in the News?
Over the past two decades, as genome sequencing technologies have become increasingly accessible, scientists have made significant strides in understanding the applications of Cell-free DNA.
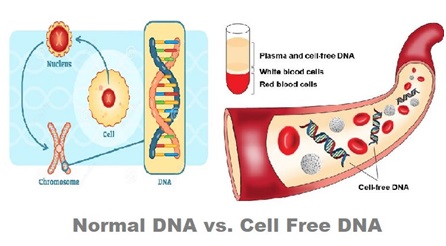
Regarding Cell-free DNA:
- In the human body, a significant portion of the DNA in the genome is safely enclosed within cells, safeguarded by specific proteins to prevent degradation.
- However, in various circumstances, certain DNA fragments are liberated from their confines and can be found outside the cells, circulating in body fluids.
- These minute fragments of nucleic acids are commonly referred to as cell-free DNA (cfDNA).
How Cell-free DNA is generated/released?
- The generation and release of Cell-free DNA (cfDNA) can occur through various mechanisms.
- One such process is when a cell undergoes cell death, leading to the degradation of nucleic acids and subsequent release of cfDNA.
- The degradation of cfDNA is influenced by a diverse set of processes, resulting in variations in the amount, size, and origin of cfDNA.
- Furthermore, this release of cfDNA can be associated with different biological processes, including those essential for normal development, the progression of certain cancers, and various other diseases.
- The generation and release of cfDNA can be triggered by a range of situations and processes, making it a versatile biomarker with potential implications in various health conditions.
Applications of Cell-free DNA (cfDNA):
- One of the most prevalent uses of cfDNA is in non-invasive prenatal testing, where it aids in screening fetuses for specific chromosomal abnormalities.
- Also, cfDNA serves as a valuable tool for comprehending human diseases and leveraging this knowledge to enhance diagnosis, monitoring, and prognosis.
- cfDNA plays a crucial role in understanding the rejection of transplanted organs by the body.
- It shows promise as a potential biomarker for various neurological disorders, including Alzheimer's disease, neuronal tumors, stroke, and traumatic brain injury.
