Junk DNA
- 10 Nov 2025
In News:
For decades, large portions of the human genome were dismissed as “junk DNA”- genetic material assumed to have little or no functional value. However, advances in genetics, genomics, and artificial intelligence are rapidly reshaping this understanding. Recent research, particularly the discovery of cancer-linked mutations in non-coding DNA, highlights how so-called junk DNA plays a crucial role in gene regulation, genome architecture, evolution, and disease.
Understanding Junk DNA
In genetics, junk DNA refers to regions of DNA that do not code for proteins. While DNA’s primary role is to provide instructions for protein synthesis, not all DNA sequences serve this function.
- In the human genome, nearly 98% of DNA is non-coding, whereas in simpler organisms like bacteria, only about 2% of DNA is non-coding.
- A part of non-coding DNA is known to have clear functions, such as producing:
- Transfer RNA (tRNA)
- Ribosomal RNA (rRNA)
- Regulatory RNAs
- However, a substantial fraction neither codes for proteins nor produces RNA, and its function remained unclear- hence the term junk DNA.
Over time, scientists have accumulated evidence that these regions are not entirely useless. Some DNA fragments that were originally non-functional have acquired functions through exaptation—a process by which structures or sequences evolve new roles not originally shaped by natural selection.
Emerging Functional Significance of Non-Coding DNA
Modern genomics has revealed that non-coding DNA plays a vital role in:
- Gene regulation (switching genes on or off),
- Chromatin organisation,
- Genome stability,
- Evolutionary innovation.
These roles become especially critical in understanding complex diseases such as cancer, where gene regulation and genome structure are often disrupted.
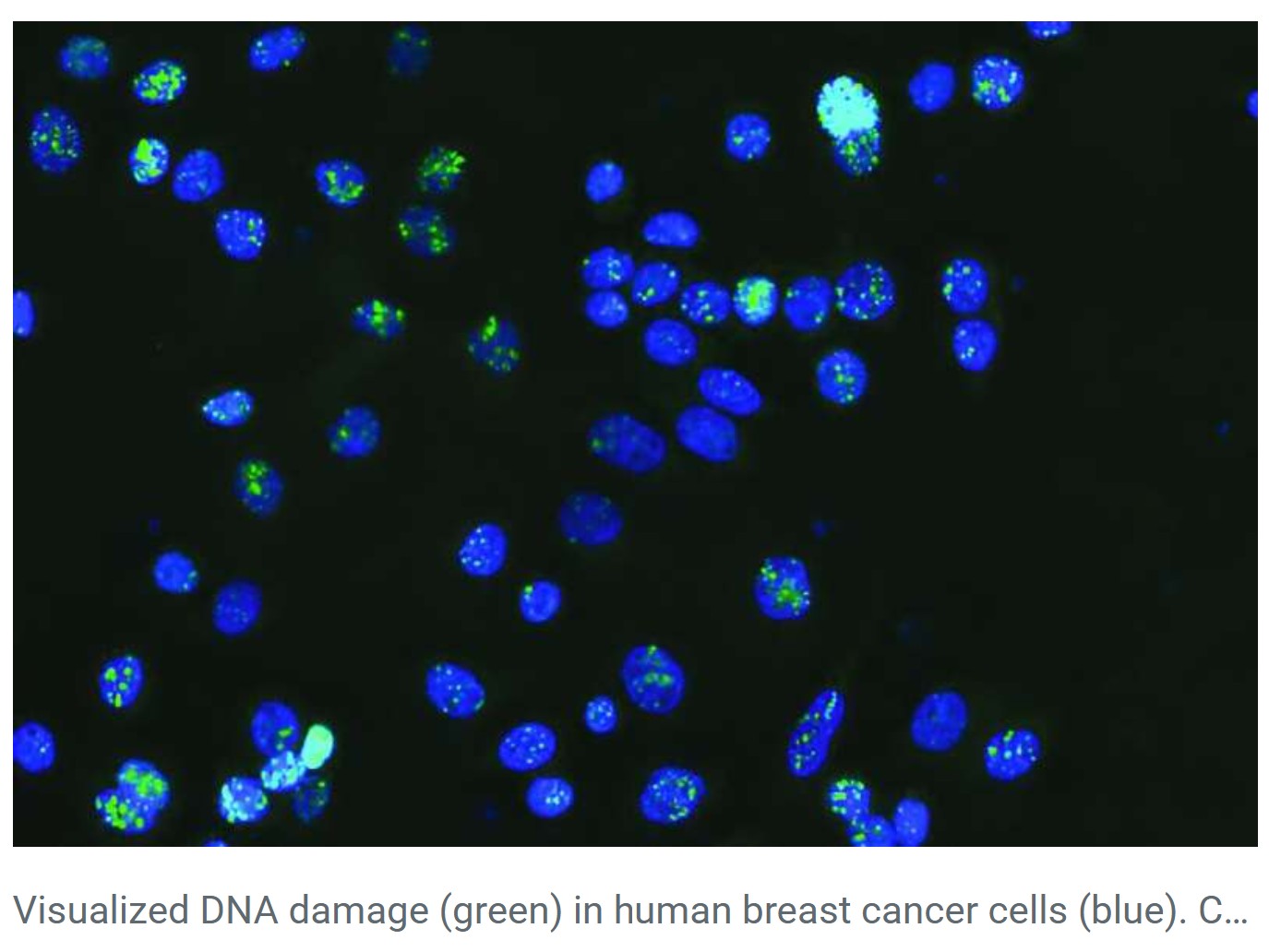
Breakthrough Discovery: Cancer Mutations in ‘Junk’ DNA
A recent study by the Garvan Institute of Medical Research, published in Nucleic Acids Research, marks a major breakthrough. Using artificial intelligence and machine learning, researchers identified a new class of cancer-driving mutations hidden in non-coding DNA.
Key Findings
- Mutations were found in non-coding regions across at least 12 cancer types, including breast, prostate, and colorectal cancers.
- Every tumour sample analysed had at least one mutation in these critical non-coding regions.
- These mutations were located at specific DNA sites that bind a protein called CTCF (CCCTC-binding factor).
Role of CTCF and Genome Architecture
CTCF is a key protein that helps fold long strands of DNA into precise three-dimensional (3D) structures inside the nucleus. These structures act as genomic “anchors”, bringing distant DNA regions together and controlling which genes are expressed.
- Some CTCF binding sites are “persistent anchors”, meaning they are present across many cell types.
- Mutations at these sites disrupt the 3D organisation of the genome, leading to abnormal gene activation or suppression.
- Such disruptions give cancer cells a survival and growth advantage, turning these sites into mutational hotspots.
To identify these sites, researchers developed an AI-based tool called CTCF-INSITE, which analysed genomic and epigenomic data from over 3,000 tumour samples using data from the International Genome Consortium.
Implications for Cancer Diagnosis and Treatment
This discovery has far-reaching implications:
- Universal cancer targets: Since the same non-coding mutations appear across multiple cancers, therapies could potentially work across cancer types rather than being mutation-specific.
- Early diagnosis: Alterations in these genomic anchors could serve as biomarkers for early cancer detection.
- New treatment strategies: Researchers plan to use CRISPR gene-editing to study how correcting these mutations affects cancer progression.
- AI in healthcare: The study demonstrates how artificial intelligence can uncover hidden patterns in vast biological data sets.
